Real-time quantitative polymerase chain reaction (RT-qPCR) is an analytical method widely used in research and diagnostics. In this method, the expression level of the target gene is calculated in relation to the reference gene/genes. It is assumed that the level of expression of these genes is constant regardless of environmental factors to which the cell is subjected or its physiological state. Choosing the appropriate reference genes is very important. Selection only based on literature, may have consequences in an incorrect estimation of the studying genes expression, and thus in the faulty interpretation of the results.
In this developed tool, the group of analyzed samples (an experimental model) is divided into several sub-models consisting of a smaller number of samples (at least two samples) and then in each of these models, the best reference gene/pair of genes is selected from the group of potential reference genes using the NormFinder algorithm. The selected gene or pairs of genes being the best reference in a given model constitute the basis for further stages of the analysis, i.e. validation of the trend of target gene expression among particular samples.
Within each model, statistical analyzes are carried out based on appropriately selected tests, showing significant statistical differences in target gene expression between the given samples across individual models. The next step is to compare the consistency of the obtained results/values including statistics for a given target gene within all tested models. The program sets the so-called coherence score describing the level of consistency of the obtained results. Achieving an unsatisfactory value of this coefficient - below the value acceptable by the researcher - leads to the next stage of analysis, i.e. removal of the potential reference gene with the weakest stability value in each model, re-selection of the reference gene/pair of genes, and re-validation of the results in all tested models.
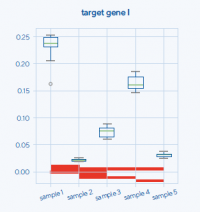
The developed tool combines the functionalities of programs selecting the best reference genes, and spreadsheet programs, allowing for an analysis of the expression level of the target gene, statistical analysis, and graphic interpretation of the results. An important element of the solution is the implementation of a specific RT-qPCR data analysis scheme (removal of the potential reference genes with the weakest stability and subsequent analysis of raw data) and identification of the coherence score value defining the reliability/credibility of the analysis. The useful improvement of the entire process is also the automatic generation of box plots presenting the obtained results of the analysis, including the statistical significance marked on the graph between the examined samples of the experimental model (Fig. 1). The program enables the simultaneous, independent analysis of many target genes.

The developed informatic tool allows:
1) selecting an improved reference with higher stability from the initial set of candidate reference genes;
2) estimating the real level of expression of the genes tested;
3) correct biological interpretation of the results based on a comprehensive analysis of raw data obtained in the RT-qPCR reaction for multi-element experimental models (large research groups consisting of cell lines, samples taken from patients or animals).
Please click below for a pamphlet.
The program is hereby made available free of charge on the basis of the so-called BSD three-clause license.
Copyright 2021 Jagiellonian University, Dr hab. Dorota Hoja-Łukowicz, prof. Jagiellonian University, Ph.D. Marcelina Janik.
1.Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
2. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
3. Neither the name of the Institute nor the names of its associates may be used to support or promote products based on this software without specific prior written consent.
Download:
GenExpA.zip
information / broker of Jagiellonian University